Evolution of biological systems
An algorithm to systemically infer the evolutionary root of orthologous groups
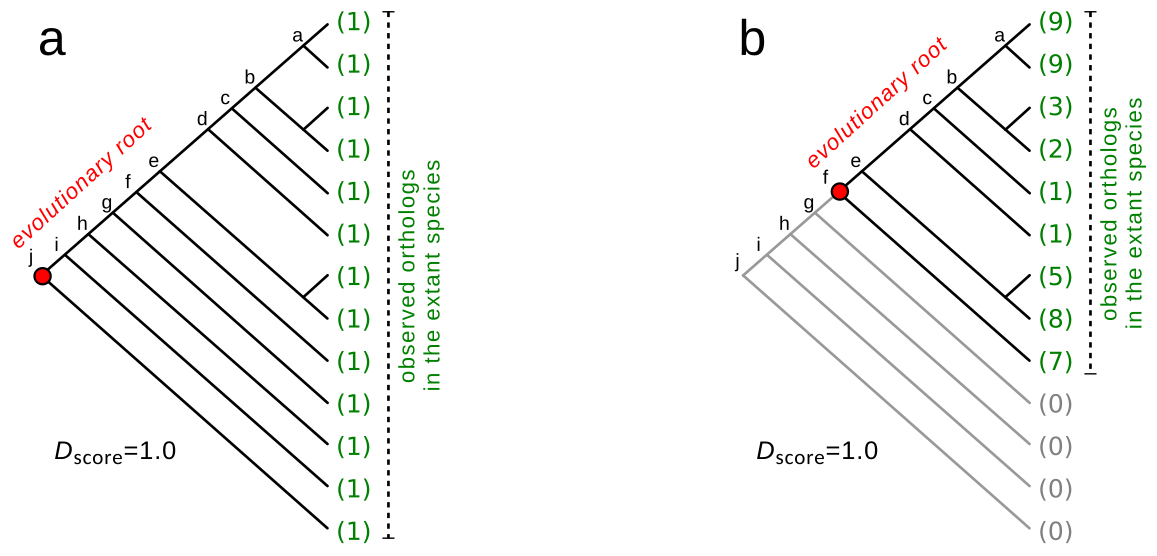
The development of sequencing technologies in the past two decades allowed the study of the phylogenetic relationship of several model organisms. An interesting question relies on the mechanisms behind how new genes arise and how they interact and evolve together to build the intricate network of genes observed in the organism’s biochemical pathways. A strategy is proposed in Castro et al, 2008 and in Dalmolin et al, 2011 papers, and it envolves two main steps. First, a consensus phylogeny for eukaryotes is constructed. The second step involves reconstructing the evolutionary scenario for each set of orthologous genes. This step aims to find the most parsimonious mapping of orthologs on the species tree, determining the earliest ortholog for each orthologous group associated with a given gene. The algorithm is implemented as an R/Bioconductor package called geneplast.
This approach was used to unravel the evolution of biochemical systems, such as…
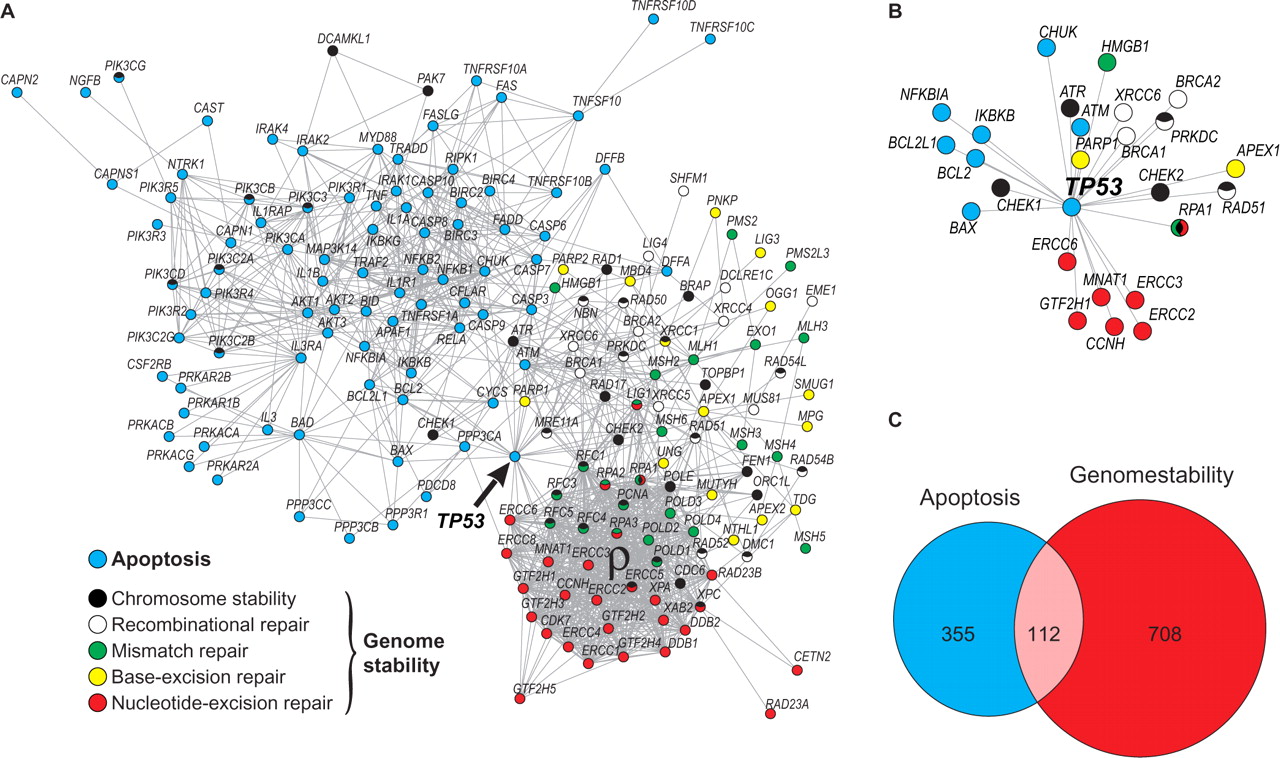
Human apoptosis and genome-stability
In the Castro et al., 2008, we estimated the evolutionary root for the apoptosis and the genome-stability genes and we found that many cancer genes are located in the earlier, more plastic and less essential region.
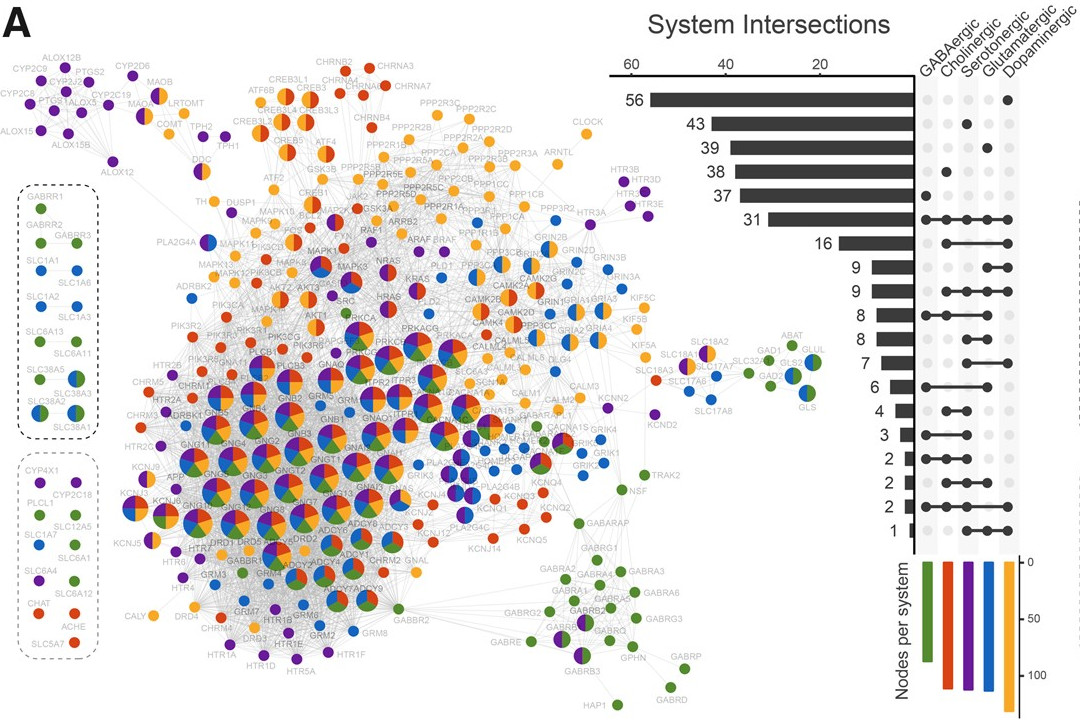
Human synapses
In the Viscardi et al. 2020 paper, we aimed to reconstruct the evolutionary history of the human neurotransmission gene network by analyzing genes in major neurotransmitter systems. The findings suggest that the emergence of receptor families, particularly ionotropic receptors, at the Human–Cnidaria Last Common Ancestor played a crucial role in the evolution of synapses, even before their establishment in Ctenophores.
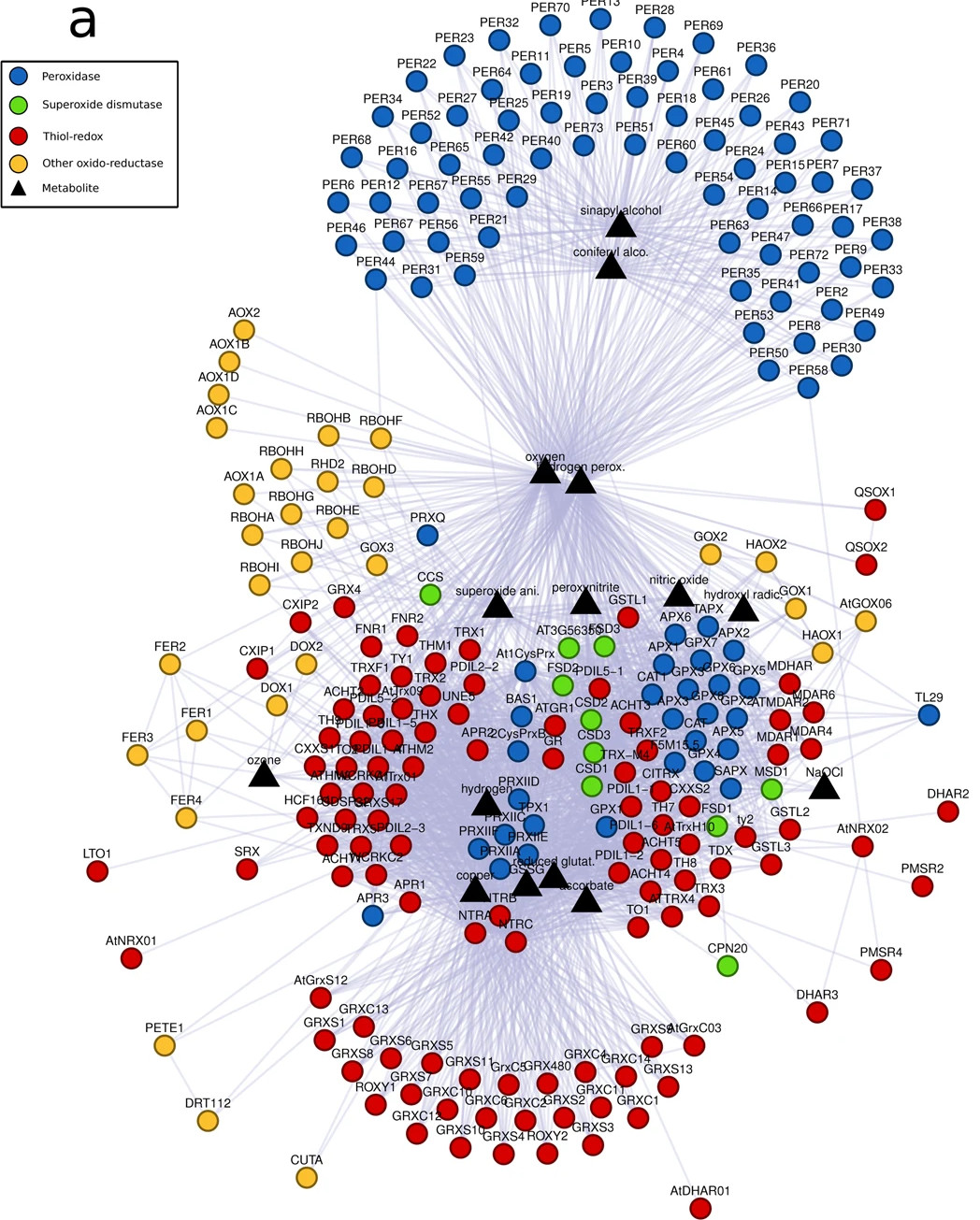
The reactive oxygen species system in Arabidopsis thaliana
In the Oliveira et a., 2019 paper, we used orthologous groups information from 238 eukaryotes to perform an evolutionary analysis of genes related to reactive oxygen species (ROS) in Arabidopsis thaliana. We found two interconnected clusters of ROS genes: one formed by SOD-related, Thiol-redox, peroxidases, and other oxido-reductase; and the other formed entirely by class III peroxidases. Each cluster emerged in different periods of evolution: the cluster formed by SOD-related, Thiol-redox, peroxidases, and other oxido-reductase emerged before opisthokonta-plant divergence; the cluster composed by class III peroxidases emerged after opisthokonta-plant divergence and therefore contained the most recent network components. According to our results, class III peroxidases are in expansion throughout plant evolution, with new orthologs emerging in each evaluated plant clade divergence.
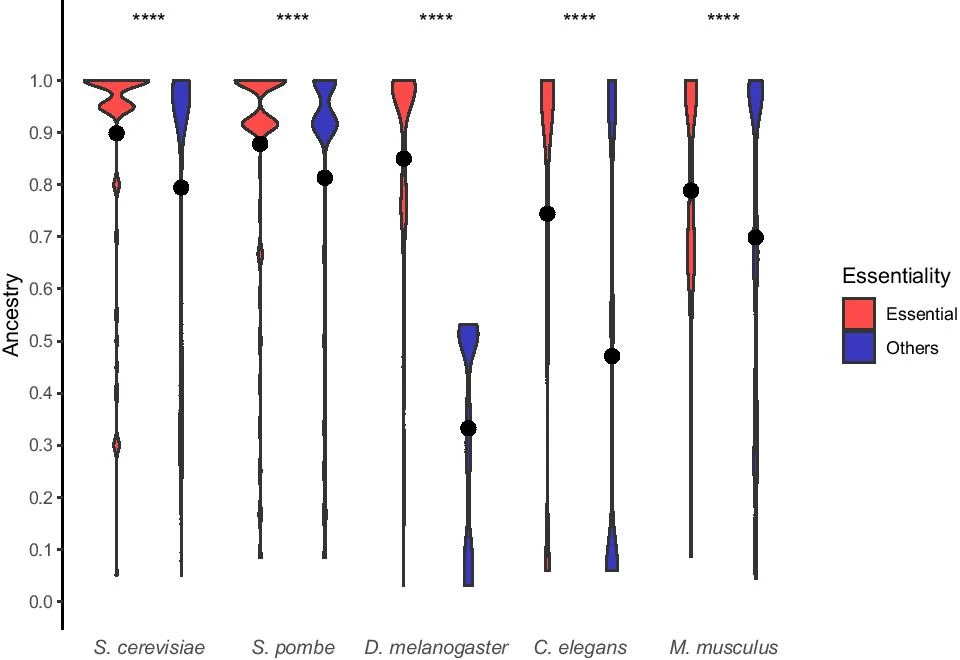
Essential genes in eukaryotes
In De Souza et al, 2021 paper, essential genes from five model eukaryotes were analyzed to determine if they differ in age from non-essential genes. The study compared network properties and investigated the biological functions of essential genes in each species, revealing that essential genes are rooted in different evolutionary periods. Despite varying origins, essential genes tend to be older and more connected than other genes across all evaluated model species.