Metagenomics analyses
Where do we start?
Metagenomics data, or data containing the full genetic content of an environmental sample, constitutes one of our current research topics at the Dalmolin Group. Metagenomics data provides a great potential for studying the diversity and functional activity of complex microbial environments.
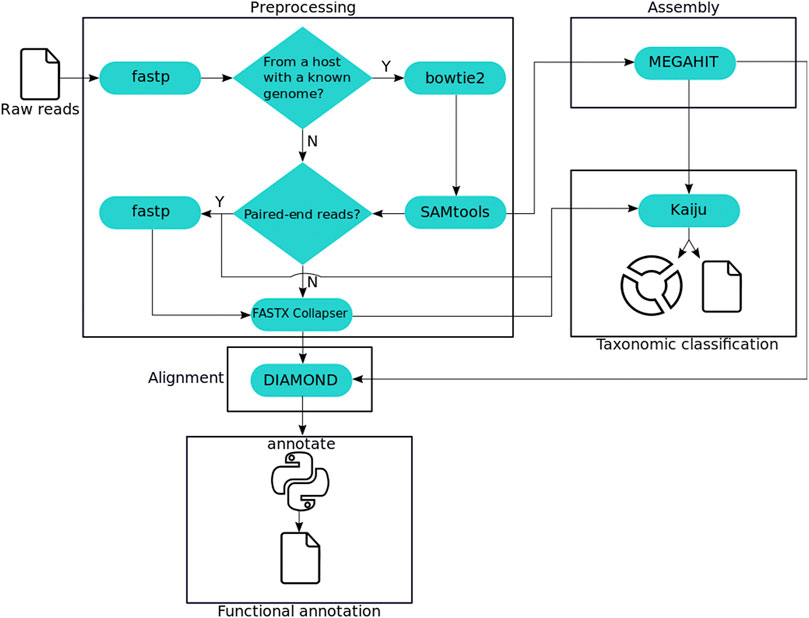
Our studies of metagenomics data began with the development of our in-house pipeline for processing these data, MEDUSA (Morais et al, 2022). With MEDUSA, we aimed to build a sensitive, flexible and reproducible pipeline that produces taxonomic classification and functional annotation of metagenomics reads. Below, you can see a diagram detailing the different steps of the original MEDUSA pipeline. We later reimplemented MEDUSA in another workflow orchestration language, Nextflow, this time being called EURYALE.
Current work and future directions
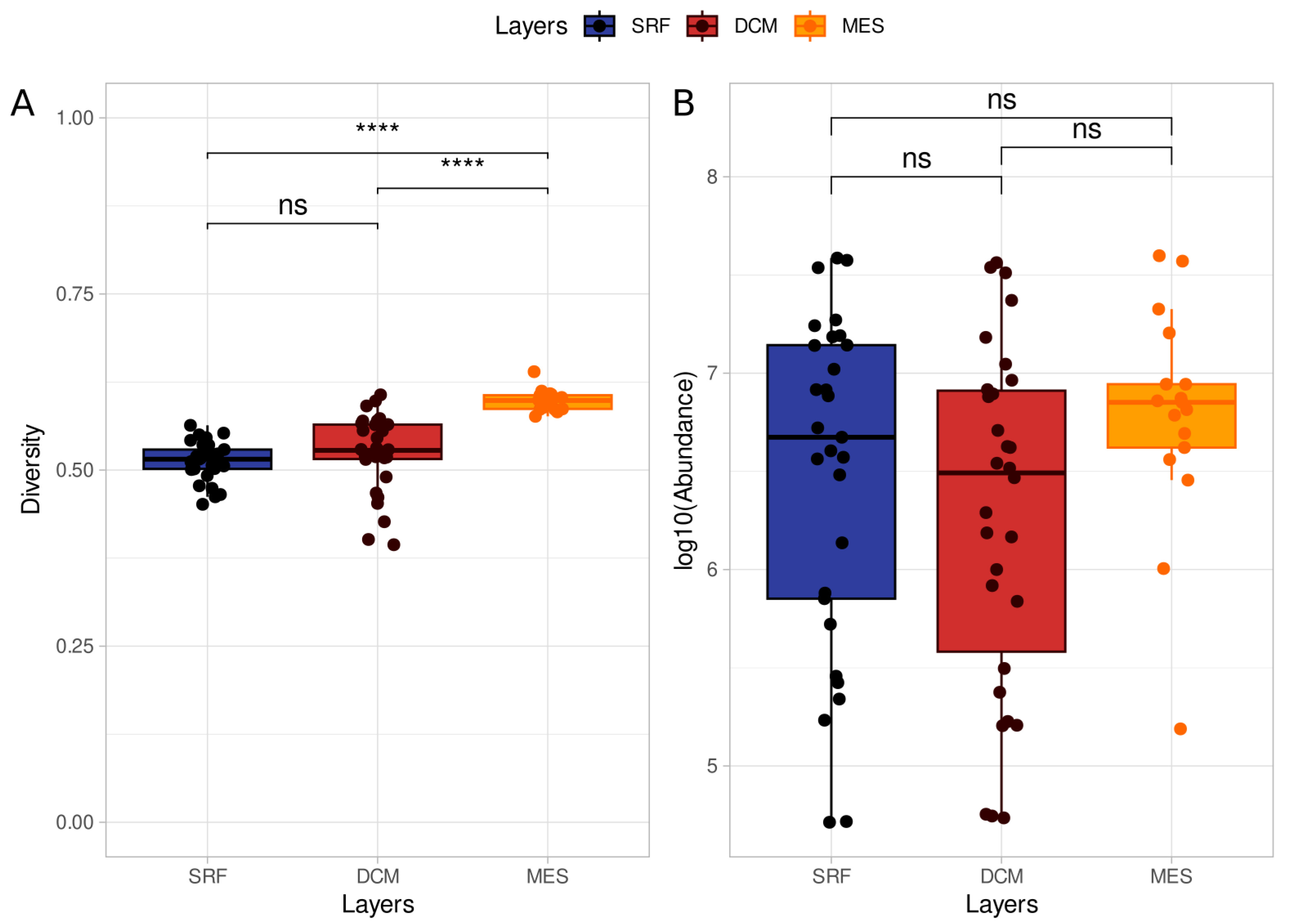
Some of our work exploring metagenomics data is highlighted by “Metagenomic Analyses Reveal the Influence of Depth Layers on Marine Biodiversity on Tropical and Subtropical Regions” (Santiago et al, 2023). In this work, using MEDUSA, we observed differences in diversity between three ocean layers in the Tropical and Subtropical regions and also noted different functional annotations between these layers.
And now, with MEDUSA/EURYALE and all of the software in the metagenomics ecosystem, we aim to keep exploring metagenomics data, with a focus on functional analysis.